Raccoon2
Raccoon2 is a graphical interface for preparing and analyzing AutoDock Vina virtual screenings.
Raccoon2 is an evolution of the previous version of Raccoon. The code has been re-written from scratch and designed in a more modular fashion, providing a much robust and flexible way of adding new features.
Features
In this first release, Raccoon2 supports only Linux Cluster computational resources with PBS and SGE schedulers. The next version (currently in development) will provide support for other computational resources like the local workstation and Opal services. Results generated outside Raccoon2 can be imported and analyzed. Also, in this release the only supported docking engine is AutoDock Vina.
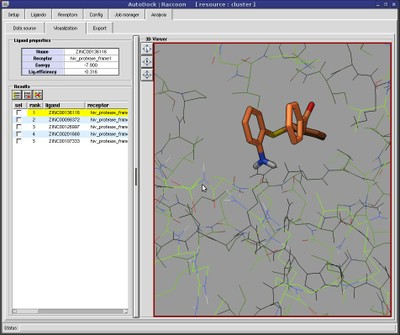

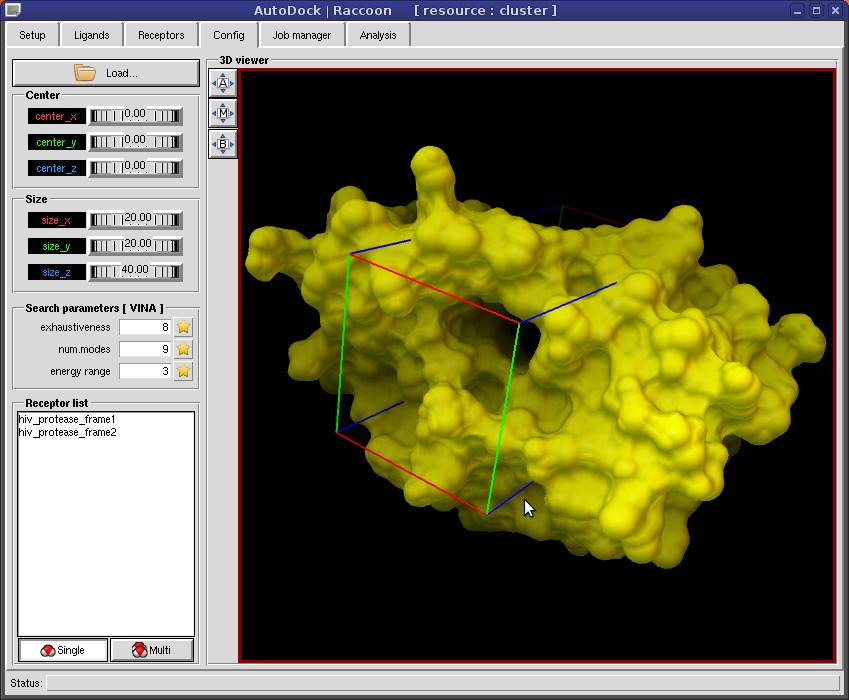
Raccoon provides features to automate common operations performed when preparing and analyzing a virtual screening:
- automatic server connection manager
- automatic installation and setup of docking services (Vina only, in v.1.0-beta)
- remote library creation interface and upload
- multiple receptor structures and flexible residues
- graphical interface for docking setup and grid box
- graphical management of remote jobs (submit, start, kill, restart)
- automatic download and pre-processing of results (i.e. extraction of energy, interactions, etc…)
- results filtering by properties (energy, ligand efficiency) and interactions
Documentation
Raccoon2 - example files (ZIP) (4101 downloads )Raccoon2 Quickstart (4782 downloads )
Installation
Raccoon2 is included in all MGLTools versions starting from v.1.5.7-rc1 on. Installers can be downloaded from here.
Beta version FAQ/limitations
- SSH connection is established using port 22; next version will provide an interface to specify custom ports
- Submission to Linux clusters are made using the user account default queue requesting 1Gb memory/24h walltime; next version will provide an interface to specify custom scheduler requests
- Only PDBQT files can be used to prepare virtual screenings. Until this functionality is added to Raccoon2, use the previous Raccoon interface to generate PDBQT files.
- This version supports only AutoDock Vina. The next version will bring support for AutoDock4.
- No local calculations are supported in the beta version
Citation and reference
Citation
When using Raccoon, please cite:
Computational protein-ligand docking and virtual drug screening with the AutoDock suite.
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ, Nature Protocols, 2016 May, doi:10.1038/nprot.2016.051
Help and support
As for all the MGL software, community and developers support for Raccoon is available through the Internet.
Any suggestions for improvements or bug reports are welcome.
Subscribe to the AutoDock mailing list as described here: https://autodocksuite.scripps.edu/support
The best way to report a problem is to write a message with the prefix “Raccoon: ” in the subject and a short description of the issue.
Any error messages and further details should be reported in the body of the email.