AutoDock Hydrated Docking protocol
NOTE: This tutorial is obsolete. Hydrated docking is now compatible with AutoDock Vina since v.1.2.0. The updated tutorials are available here.
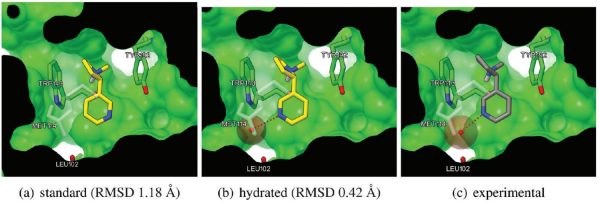
The package contains scripts, data files and example structures to perform an AutoDock hydrated docking as described in the paper “A Force Field with Discrete Displaceable Waters and Desolvation Entropy for Hydrated Ligand Docking”, J.Med.Chem. 2012
This implementation requires some degree of familiarity with the standard AutoDock protocol and with the command-line interface.
Be aware that with this implementation of the method, it is difficult to compare results obtained with very diverse ligands without doing extra of post-processing on the results, because the energy estimation needs to be normalized. For this reason, the method is not suitable for virtual screenings. This doesn’t affect the structural accuracy, so comparisons within docking poses are fine. An improved scoring function to overcome this issue is in the works.
For questions and help, please use the AutoDock mailing list